Multinomial Logistic Regression with Heavy-Tailed Priors
Longhai Li and Steven Liu
2025-12-14
Source:vignettes/simu.Rmd
simu.RmdData Generation
Load the necessary libraries:
library(HTLR)
library(bayesplot)
#> This is bayesplot version 1.15.0
#> - Online documentation and vignettes at mc-stan.org/bayesplot
#> - bayesplot theme set to bayesplot::theme_default()
#> * Does _not_ affect other ggplot2 plots
#> * See ?bayesplot_theme_set for details on theme settingThe description of the dataset generating scheme is found from Li and Yao (2018).
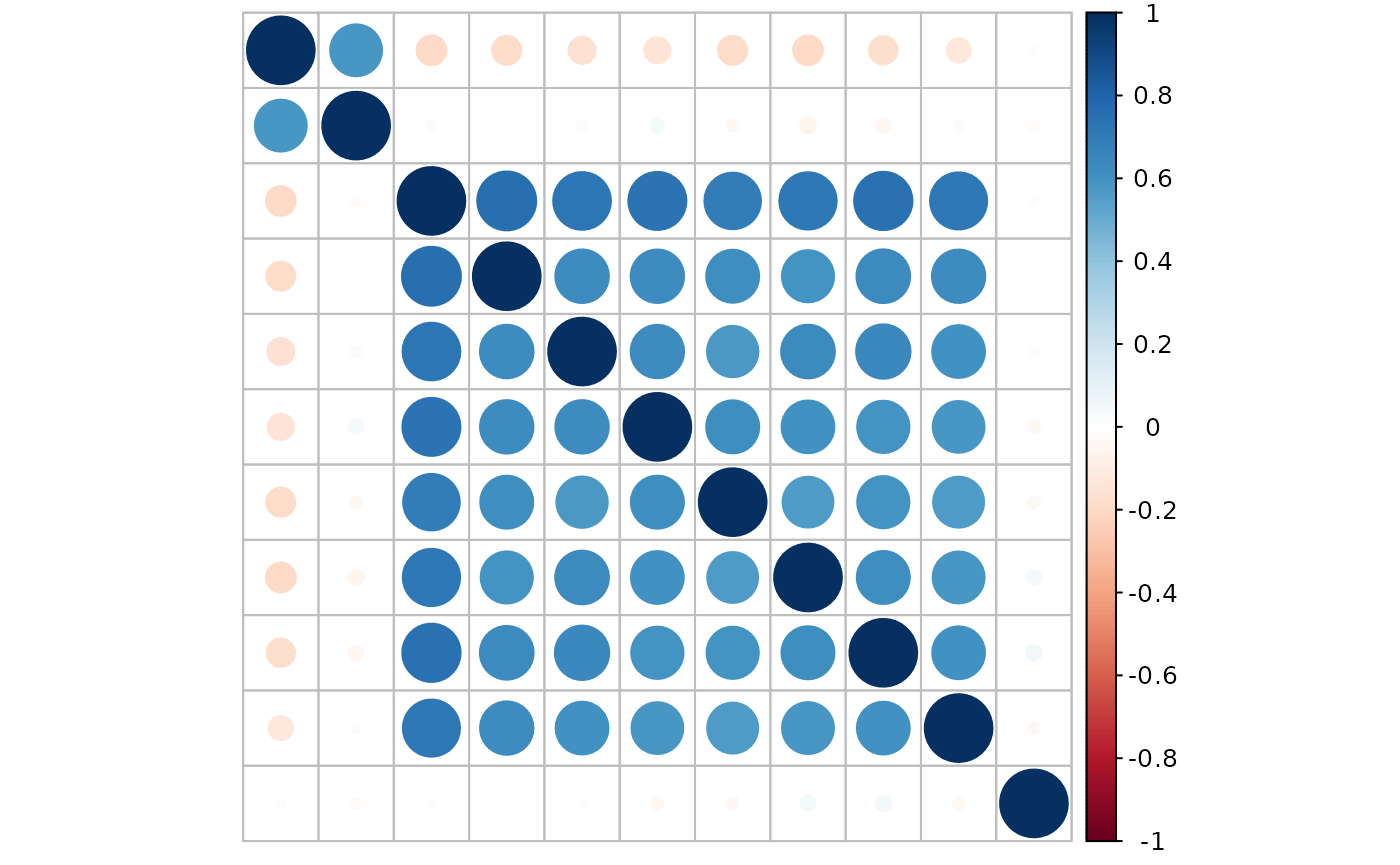
There are 4 groups of features:
feature #1: marginally related feature
feature #2: marginally unrelated feature, but feature #2 is correlated with feature #1
feature #3 - #10: marginally related features and also internally correlated
feature #11 - #2000: noise features without relationship with the y
SEED <- 123
n <- 510
p <- 2000
means <- rbind(
c(0, 1, 0),
c(0, 0, 0),
c(0, 0, 1),
c(0, 0, 1),
c(0, 0, 1),
c(0, 0, 1),
c(0, 0, 1),
c(0, 0, 1),
c(0, 0, 1),
c(0, 0, 1)
) * 2
means <- rbind(means, matrix(0, p - 10, 3))
A <- diag(1, p)
A[1:10, 1:3] <-
rbind(
c(1, 0, 0),
c(2, 1, 0),
c(0, 0, 1),
c(0, 0, 1),
c(0, 0, 1),
c(0, 0, 1),
c(0, 0, 1),
c(0, 0, 1),
c(0, 0, 1),
c(0, 0, 1)
)
set.seed(SEED)
dat <- gendata_FAM(n, means, A, sd_g = 0.5, stdx = TRUE)
str(dat)
#> List of 4
#> $ X : num [1:510, 1:2000] -0.684 0.912 -0.997 -1.262 0.613 ...
#> ..- attr(*, "dimnames")=List of 2
#> .. ..$ : NULL
#> .. ..$ : chr [1:2000] "V1" "V2" "V3" "V4" ...
#> $ muj: num [1:2000, 1:3] -0.456 0 -0.456 -0.376 -0.376 ...
#> $ SGM: num [1:2000, 1:2000] 0.584 0.597 0 0 0 ...
#> $ y : int [1:510] 1 2 3 1 2 3 1 2 3 1 ...Look at the correlation between features:
Split the data into training and testing sets:
set.seed(SEED)
dat <- split_data(dat$X, dat$y, n.train = 500)
str(dat)
#> List of 4
#> $ x.tr: num [1:500, 1:2000] -0.903 -0.632 1.111 1.446 -0.43 ...
#> ..- attr(*, "dimnames")=List of 2
#> .. ..$ : NULL
#> .. ..$ : chr [1:2000] "V1" "V2" "V3" "V4" ...
#> $ y.tr: int [1:500] 1 1 2 2 3 3 3 1 2 1 ...
#> $ x.te: num [1:10, 1:2000] 1.955 1.188 -0.942 -1.387 0.879 ...
#> ..- attr(*, "dimnames")=List of 2
#> .. ..$ : NULL
#> .. ..$ : chr [1:2000] "V1" "V2" "V3" "V4" ...
#> $ y.te: int [1:10] 2 2 3 3 2 3 1 2 2 2Model Fitting
Fit a HTLR model with all default settings:
set.seed(SEED)
system.time(
fit.t <- htlr(dat$x.tr, dat$y.tr)
)
#> user system elapsed
#> 240.234 0.164 60.703
print(fit.t)
#> Fitted HTLR model
#>
#> Data:
#>
#> response: 3-class
#> observations: 500
#> predictors: 2001 (w/ intercept)
#> standardised: TRUE
#>
#> Model:
#>
#> prior dist: t (df = 1, log(w) = -10.0)
#> init state: lasso
#> burn-in: 1000
#> sample: 1000 (posterior sample size)
#>
#> Estimates:
#>
#> model size: 4 (w/ intercept)
#> coefficients: see help('summary.htlr.fit')With another configuration:
set.seed(SEED)
system.time(
fit.t2 <- htlr(X = dat$x.tr, y = dat$y.tr,
prior = htlr_prior("t", df = 1, logw = -20, sigmab0 = 1500),
iter = 4000, init = "bcbc", keep.warmup.hist = T)
)
#> user system elapsed
#> 371.925 0.849 94.654
print(fit.t2)
#> Fitted HTLR model
#>
#> Data:
#>
#> response: 3-class
#> observations: 500
#> predictors: 2001 (w/ intercept)
#> standardised: TRUE
#>
#> Model:
#>
#> prior dist: t (df = 1, log(w) = -20.0)
#> init state: bcbc
#> burn-in: 2000
#> sample: 2000 (posterior sample size)
#>
#> Estimates:
#>
#> model size: 4 (w/ intercept)
#> coefficients: see help('summary.htlr.fit')Model Inspection
Look at the point summaries of posterior of selected parameters:
summary(fit.t2, features = c(1:10, 100, 200, 1000, 2000), method = median)
#> class 2 class 3
#> Intercept -2.7354013973 -1.119076e+00
#> V1 8.2082272603 -6.028105e-01
#> V2 -4.5701096071 2.313064e-01
#> V3 -0.9363523582 3.214379e+00
#> V4 0.0009046690 -2.152431e-03
#> V5 -0.0051665924 -5.631749e-05
#> V6 -0.0072774286 1.654051e-03
#> V7 -0.0013794333 -2.153372e-03
#> V8 -0.0047661861 -7.569987e-03
#> V9 -0.0060317708 2.927544e-04
#> V10 -0.0005934682 1.050192e-02
#> V100 -0.0039015133 1.115288e-02
#> V200 -0.0066909487 6.128469e-04
#> V1000 0.0051105291 6.717532e-03
#> V2000 -0.0058668522 -7.001962e-03
#> attr(,"stats")
#> [1] "median"Plot interval estimates from posterior draws using bayesplot:
post.t <- as.matrix(fit.t2, k = 2)
## signal parameters
mcmc_intervals(post.t, pars = c("Intercept", "V1", "V2", "V3", "V1000"))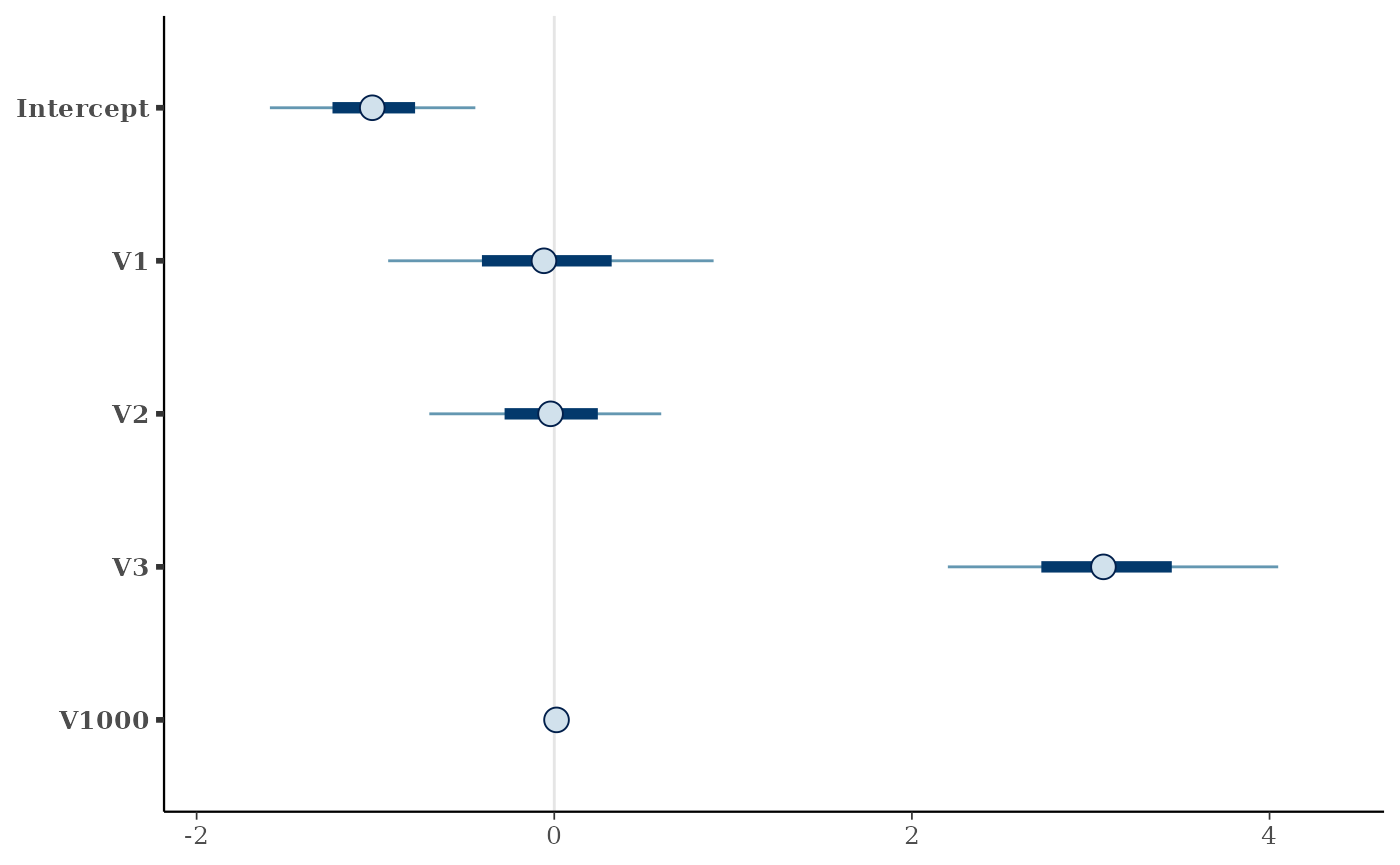
Trace plot of MCMC draws:
as.matrix(fit.t2, k = 2, include.warmup = T) %>%
mcmc_trace(c("V1", "V1000"), facet_args = list("nrow" = 2), n_warmup = 2000)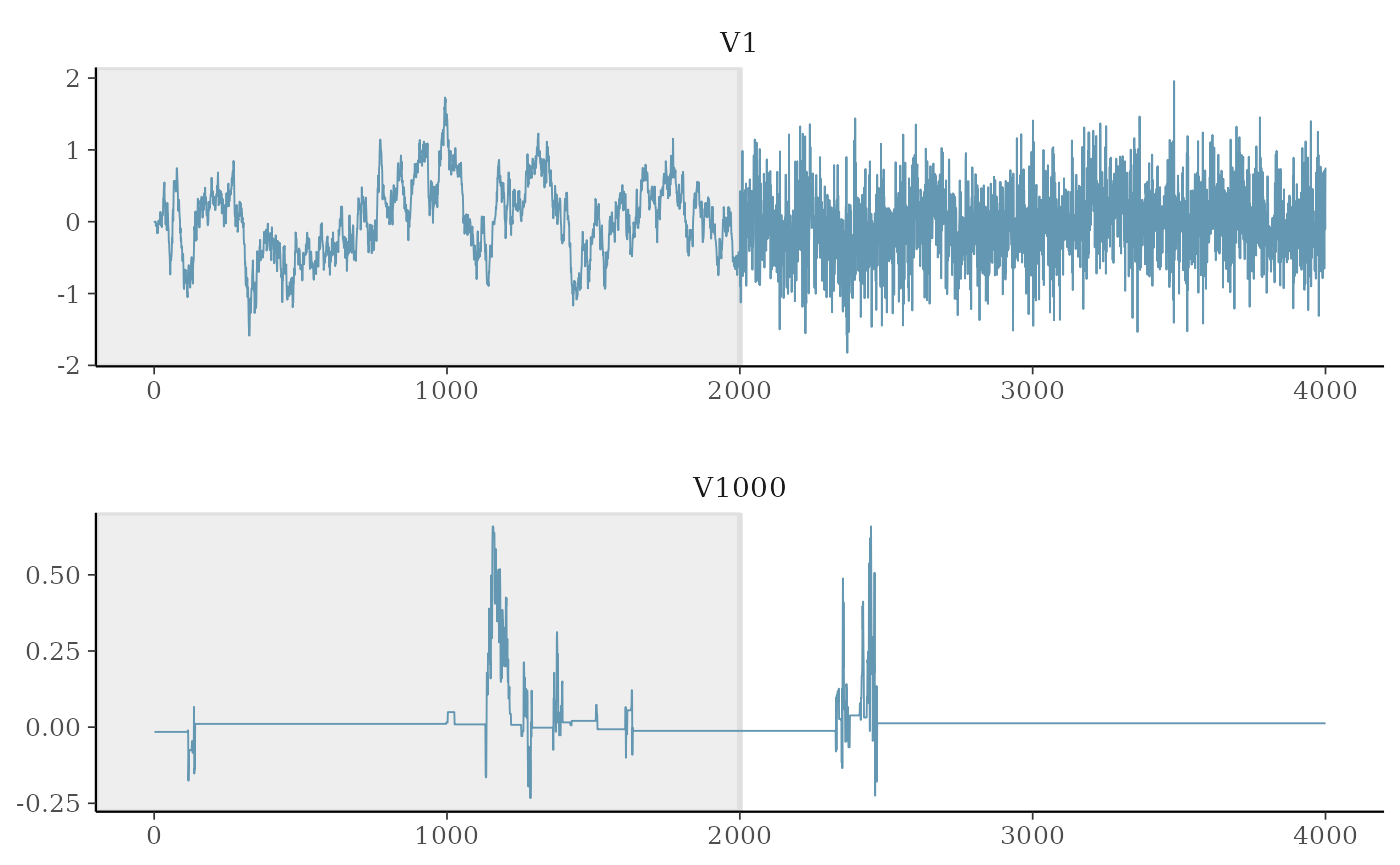
The coefficient of unrelated features (noise) are not updated during some iterations due to restricted Gibbs sampling Li and Yao (2018), hence the computational cost is greatly reduced.
Make Predictions
A glance at the prediction accuracy:
y.class <- predict(fit.t, dat$x.te, type = "class")
y.class
#> y.pred
#> [1,] 2
#> [2,] 2
#> [3,] 3
#> [4,] 3
#> [5,] 2
#> [6,] 3
#> [7,] 3
#> [8,] 2
#> [9,] 2
#> [10,] 2
print(paste0("prediction accuracy of model 1 = ",
sum(y.class == dat$y.te) / length(y.class)))
#> [1] "prediction accuracy of model 1 = 0.9"
y.class2 <- predict(fit.t2, dat$x.te, type = "class")
print(paste0("prediction accuracy of model 2 = ",
sum(y.class2 == dat$y.te) / length(y.class)))
#> [1] "prediction accuracy of model 2 = 0.9"More details about the prediction result:
predict(fit.t, dat$x.te, type = "response") %>%
evaluate_pred(y.true = dat$y.te)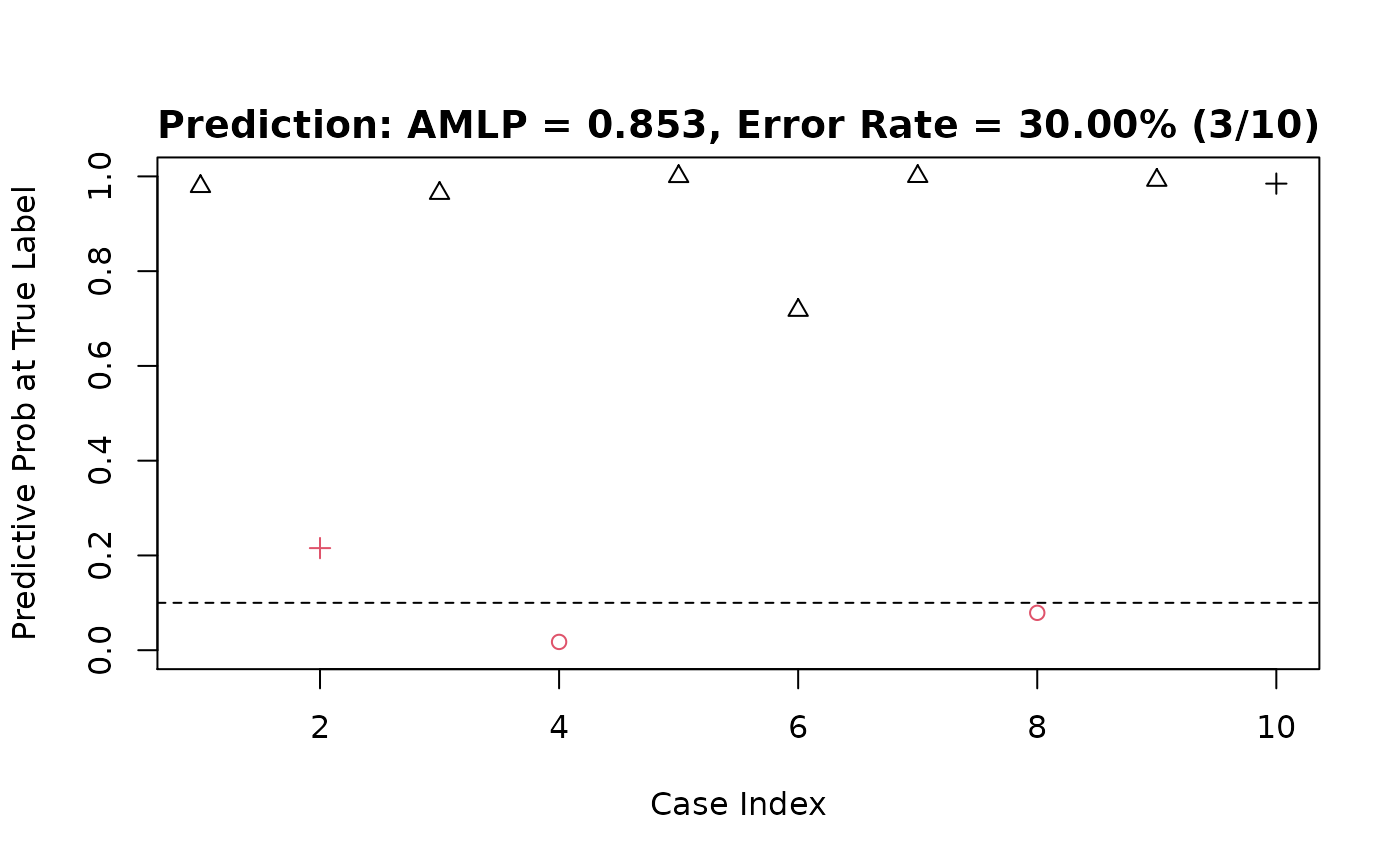
#> $prob_at_truelabels
#> [1] 0.9993980 0.9996598 0.9960531 0.9146809 0.5601952 0.4681349 0.1604013
#> [8] 0.9999976 0.9986511 0.9655200
#>
#> $table_eval
#> Case ID True Label Pred. Prob 1 Pred. Prob 2 Pred. Prob 3 Wrong?
#> 1 1 2 2.230163e-04 9.993980e-01 3.789681e-04 0
#> 2 2 2 3.397451e-04 9.996598e-01 4.055238e-07 0
#> 3 3 3 3.946910e-03 3.274933e-09 9.960531e-01 0
#> 4 4 3 8.531913e-02 1.201749e-08 9.146809e-01 0
#> 5 5 2 4.363766e-01 5.601952e-01 3.428135e-03 0
#> 6 6 3 1.231557e-01 4.087093e-01 4.681349e-01 0
#> 7 7 1 1.604013e-01 9.352412e-04 8.386635e-01 1
#> 8 8 2 2.383022e-06 9.999976e-01 3.584158e-08 0
#> 9 9 2 1.339940e-03 9.986511e-01 8.967895e-06 0
#> 10 10 2 3.263762e-02 9.655200e-01 1.842378e-03 0
#>
#> $amlp
#> [1] 0.3299063
#>
#> $err_rate
#> [1] 0.1
#>
#> $which.wrong
#> [1] 7